Access Unit is the fundamental structure allowing random and selective access to genomic data.
Genomic records, are compressed in MPEG-G. Thus, in order to search them, filter, and access, all structures of the MPEG-G shown on Figure 1, contain metadata, with information on genomic data contained in that structure, and parameters that were used to encode them.
Access Unit is the fundamental structure allowing random access, and filtering of genomic records. Each access unit, contains genomic records from only one Data Class (P, N, M, I, HM and U, Table 1), and each access unit can be search/filtered based on the positions in the reference genome (start position, end position). Furthermore, access units can be filtered using aggregated features of genomic records encapsulated in each of them, such as read number, alignment/nucleotide quality, Number of sliced reads (Table 2), that can be accessed without decompressing the data. Similarly to Datasets and datasets groups, all access units are secured independently. Thus, one may select which regions of the genome could be made available to certain users, in order to avoid accidental findings and data lick-age. Finally, in case new sequence data would be added to a given datasets, these will be added as new access units, thus reducing the time required for file compression.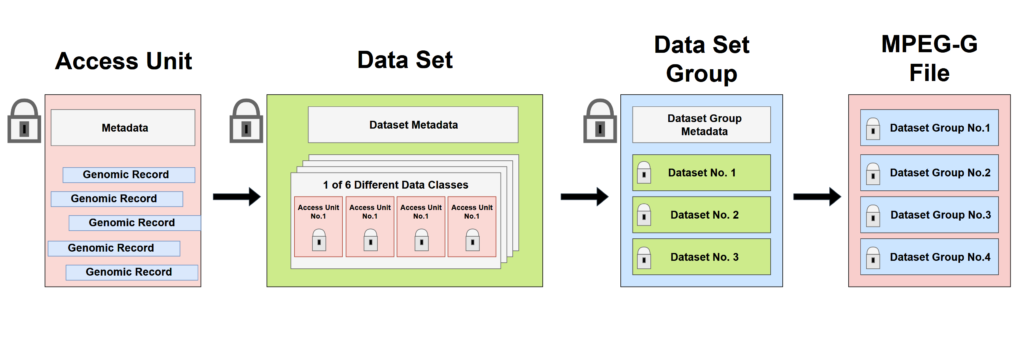 Figure 1. Key elements of MPEG-G File Format. Each Access Unit contains Genomic record belonging to only one Data Class
Figure 1. Key elements of MPEG-G File Format. Each Access Unit contains Genomic record belonging to only one Data Class
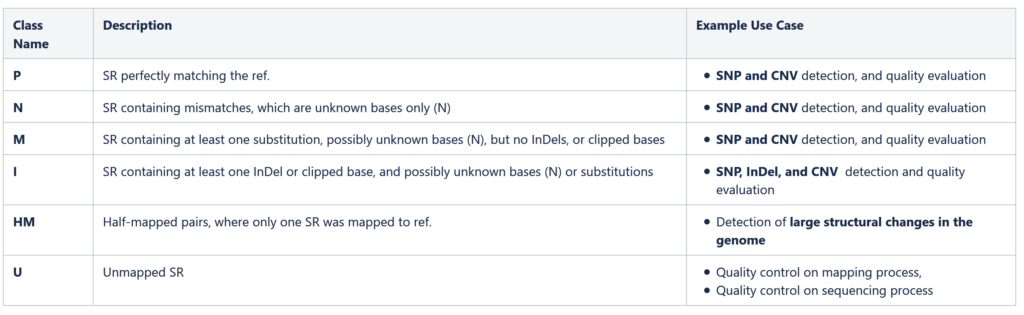
Table 1. Six different data classes defined in MPEG-G, SR – Sequence Reads, ref. – reference genome, InDel – Insertion or deletion