Part 1: Transport and Storage of Genomic Information
MPEG-G specifies a digital container format for transmission and storage of the genomic data compressed according to Part 2 of the standard. In MPEG jargon the container format used for the transport of packetized data (i.e., stream) on a telecommunication network is referred to as Transport Format, while the container format used for storage on a physical medium (i.e., file) is referred to as File Format. The process of converting a stream to a file and vice versa is normative and specified in the standard.
File Format
An MPEG-G file is organized in a file header and one or more containers named Dataset Groups. Each Dataset Group contains a Dataset Group header, optional metadata containers and encapsulates one or more Datasets. Each Dataset has a Dataset Header, optional metadata containers and carries one or more Access Units. The Access Unit is the actual container of the compressed genomic data. It includes an Access Unit header which provides a description of the compressed content (type of data, number of reads, genomic region including the compressed reads, etc.). In the case where an MPEG-G file was constructed without Descriptor Streams, the Access Unit contains a collection of Blocks of coded information that can be decoded independently using global data at the Dataset level and eventually information contained in other Access Units, such as Access Units containing data of an MPEG-G encoded reference sequence. Otherwise, the blocks of coded information are grouped by type, concatenated and stored as Descriptor Streams. The index mechanism then allows to associate a given Access Unit to the corresponding collection of Blocks. In either case, each Block is compressed using the entropy coding techniques most suitable to the measured statistical properties. This nested data structure is depicted in Figure 2. 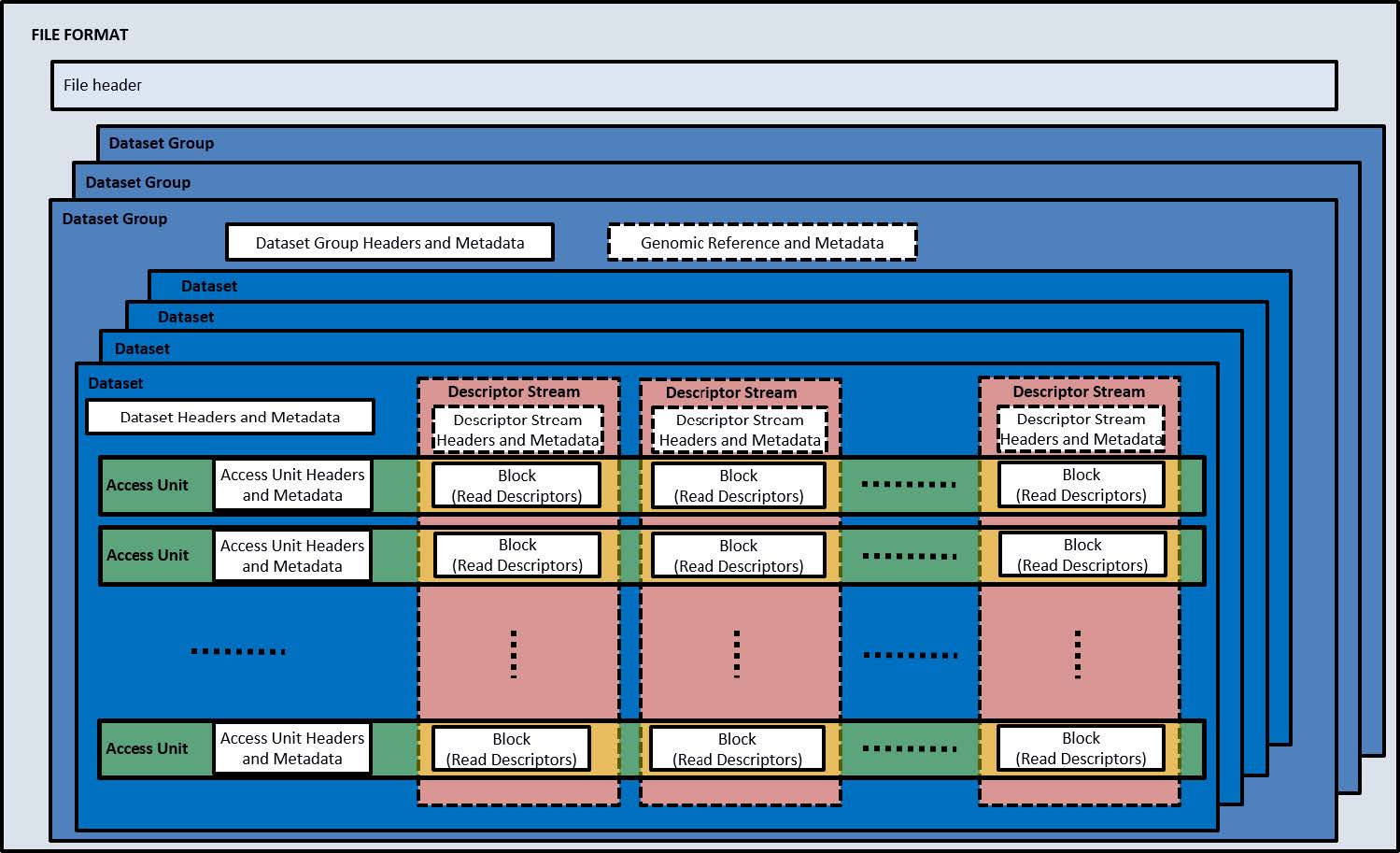 Figure 2: Key elements of the MPEG-G File Format. Multiple Dataset Groups contain multiple Datasets of sequencing data. Each Dataset is composed of Access Units containing Genomic Records pertaining to one specific Data Class. Each Access Unit is composed by Blocks of Read Descriptors.
Figure 2: Key elements of the MPEG-G File Format. Multiple Dataset Groups contain multiple Datasets of sequencing data. Each Dataset is composed of Access Units containing Genomic Records pertaining to one specific Data Class. Each Access Unit is composed by Blocks of Read Descriptors.
Transport Format
In addition to the data containers defined for the File Format, MPEG-G specifies data structures supporting packetized data transport over a network. Such structures are defined both to carry the compressed genomic data and to update metadata describing the streamed content. An example of the latter type of data is indexing information used by the receiving end to enable selective access even on partially transmitted content. The Transport Format structures are instrumental in the specification of the normative process of conversion between Transport Format (i.e., an MPEG-G stream transmitted over the internet) and File Format (i.e., an MPEG-G file stored on disk).


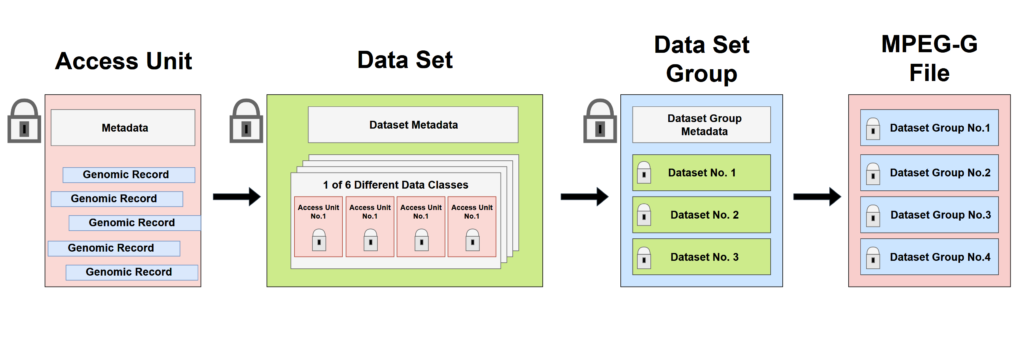 Figure 1. Key elements of MPEG-G File Format. Each Access Unit contains Genomic record belonging to only one Data Class
Figure 1. Key elements of MPEG-G File Format. Each Access Unit contains Genomic record belonging to only one Data Class